A team of researchers from Switzerland, Costa Rica and Germany has conducted a study showing which animal species could potentially serve as spillback reservoirs of severe acute repertory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
Using an in vitro airway epithelial cell (AEC) culture repository composed of various domestic, livestock, and wild animal species, the team found that SARS-CoV-2 only replicated efficiently in cultures grown from two species - the rhesus monkey and the cat.
“This is the first study employing an in vitro AEC culture repository composed of various domestic and wildlife animal species to assess the potential intermediate and spillback host reservoir spectrum of SARS-CoV-2,” say the researchers.
Whole-genome sequencing found no evidence to suggest that the predominant SARS-CoV-2 strain currently circulating the globe (the D614G variant) had undergone mutational adaptation to be able to infect the rhesus macaque and cat.
“This highlights that the currently circulating SARS-CoV-2 D614G variant can productively infect rhesus macaque and cat airway epithelial cells,” write Ronald Dijkman from the University of Bern and colleagues from Justus Liebig University Giessen, the Berlin Institute of Health and the University of Costa Rica.
Close surveillance of cats, monkeys and closely-related species is warranted to understand these species as potential spillback reservoirs for the virus, they say.
A preprint version of the paper is available on the server bioRxiv*, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The potential for human-to-animal spillover events
Although humans are currently considered the main source of SARS-CoV-2 transmission, the zoonotic origin, intermediate host species and potential spillback host reservoirs are not yet known.
Several studies have indicated that SARS-CoV-2 can spillover from humans to other animal species.
“These events are likely driven by close human-animal interactions and the conservation of crucial receptor binding motif (RBM) residues in the angiotensin-converting enzyme 2 (ACE2) orthologues, potentially facilitating SARS-CoV-2 entry,” suggest Dijkman and colleagues.
The RBM is the main region within the receptor-binding domain of SARS-COV-2 that recognizes and attaches to the human host cell receptor ACE2.
“This highlights the need to assess the potential host spectrum for SARS-CoV-2 in order to support current pandemic mitigation strategies,” says the team.
Historically, such experiments have been limited regarding the availability and diversity of animal models, housing facilities, and ethical approval, with socioemotional ethical constraints posing a particular problem in the case of companion animals and non-human primates.
Testing susceptibility to SARS-CoV-2 in 12 animal species
Now, Dijkman and team have assessed SARS-CoV-2 Susceptibility in 12 mammalian species by reproducing the initial stages of infection in well-differentiated AEC cultures reconstituted from tracheobronchial epithelial tissue taken from the animals.
By using this controlled in vitro model, the team could comply with the “reduction, replacement, refinement” (3R) principle that is applied in animal studies, while circumventing ethical and experimental constraints.
The researchers obtained post-mortem tracheobronchial airway tissue from companion animals (cat, dog), candidate animal models (rhesus macaque, ferret, and rabbit), livestock (pig, cattle, goat, llama, camel) and two bat species.
Well-differentiated AEC cultures were established for each species and then inoculated with a SARS-CoV-2 isolate (SARS-CoV-2/München-1.1/2020/929) that represents the currently circulating D614G variant.
The virus only replicated efficiently in the rhesus macaque and cat
The researchers found that only tracheobronchial cells from the rhesus macaque and cat supported efficient replication of SARS-CoV-2.
The team observed a progressive 4-log fold increase in viral RNA load at 72- and 96-hours post-infection in the rhesus macaque and cat AEC cultures, while AEC cultures from the remaining species showed either a continuous or decreasing viral RNA load.
To test whether this observed Susceptibility to SARS-CoV-2 corresponds to the amino acid sequence conservation of the RBM in ACE2, the team conducted an in silico analysis of the available ACE2 protein sequences.
This revealed that, compared with humans, the RBM regions that interact with the receptor-RBD of SARS-CoV-2 are well conserved in both the rhesus macaque and the cat, whereas they were more diverse in other species.
SARS-CoV-2 had not needed to adapt in order to infect the animals
Studies have previously demonstrated that SARS-CoV-2 can undergo rapid genetic changes in vitro.
Given the observed viral replication in the rhesus macaque and cat AEC cultures, the researchers performed whole-genome sequencing to assess whether any mutations suggestive of viral adaptation had occurred.
Neither the viral inoculum nor the progeny virus showed any obvious signs of the nucleotide transitions that lead to nonsynonymous mutations, irrespective of the animal species.
“Whole-genome sequencing indicated that the current circulating SARS-CoV-2 D614G-variant can efficiently infect rhesus macaque and cat airway epithelial cells,” writes the team.
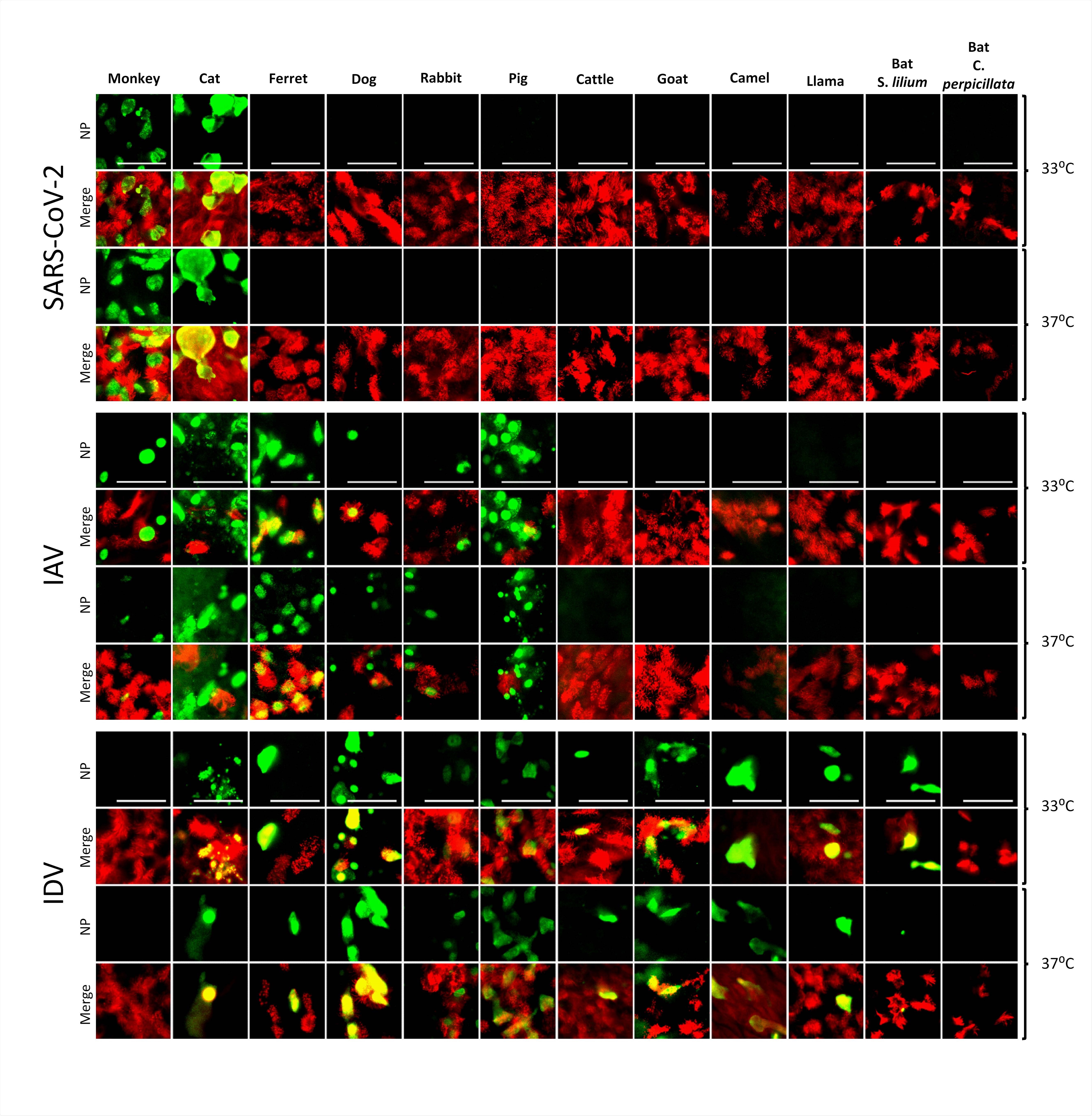
Tropisms of SARS-CoV-2, IAV, and IDV in infected AEC cultures from diverse mammalian species. Well-differentiated animal AEC cultures were inoculated with either 30.000 pfu of SARS-CoV- 2 (SARS-CoV-2/München-1.1/2020/929), 10.000 TCID50 of IAV/Hamburg/4/2009 (H1N1pdm09) or IDV (D/bovine/Oklahoma/660/2013). Virus-infected AEC cultures were incubated at respectively 33°C or 37°C and fixed 96 hpi (for SARS-CoV-2) or 48 hpi (for IAV and IDV). Following fixation, virus-infected cultures were stained using antibodies against either SARS-CoV-2, IAV, or IDV Nucleocapsid protein (green), and ß-tubulin (cilia, red). Images were acquired using an EVOS FL Auto 2 Imaging System equipped with a 40x air objective. Scale bar = 50 µm.
The researchers recommend close surveillance of the rhesus macaque and cat
The team says that these findings, together with previously documented spillover events, suggest that close surveillance of these animals, including closely-related species in the wild, in captivity and in household settings is warranted.
“Taken together, our results highlight that in vitro well-differentiated airway epithelial models in combination with high throughput genomic analysis can be applied as viable surrogate models to refine, reduce, and replace animal experimentation to evaluate host tropism of respiratory viruses, and thereby providing important insight into the host spectrum of SARS-CoV-2,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Dijkman R, et al. Susceptibility of well-differentiated airway epithelial cell cultures from domestic and wildlife animals to SARS-CoV-2. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.11.10.374587, https://www.biorxiv.org/content/10.1101/2020.11.10.374587v1
- Peer reviewed and published scientific report.
Gultom, Mitra, Matthias Licheri, Laura Laloli, Manon Wider, Marina Strässle, Philip V’kovski, Silvio Steiner, et al. 2021. “Susceptibility of Well-Differentiated Airway Epithelial Cell Cultures from Domestic and Wild Animals to Severe Acute Respiratory Syndrome Coronavirus 2.” Emerging Infectious Diseases 27 (7): 1811–20. https://doi.org/10.3201/eid2707.204660. https://wwwnc.cdc.gov/eid/article/27/7/20-4660_article.
Article Revisions
- Mar 31 2023 - The preprint preliminary research paper that this article was based upon was accepted for publication in a peer-reviewed Scientific Journal. This article was edited accordingly to include a link to the final peer-reviewed paper, now shown in the sources section.